Open GaussView and click File->Open…, then open a checkpoint (.chk) files. After the GaussView image appears like below, click from the toolbar Results->Surfaces/Contours..
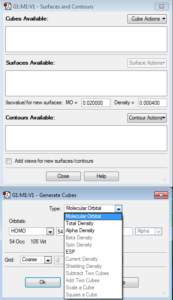
A new window will open. Select the Cube Actions dropdown and click New Cube. Make sure that the top “Type” dropdown is set to Molecular Orbital. Here, you can choose what type of molecular orbital you would like to illustrate (under the HOMO dropdown). For example, the options include HOMO, LUMO, Occupied, Virtual, or you can choose your own combination or molecular orbitals by number. Once you choose, click OK. Then, click the Surface Actions dropdown and click New Surface. This will add the MO’s to your GaussView image.
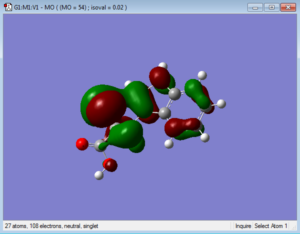
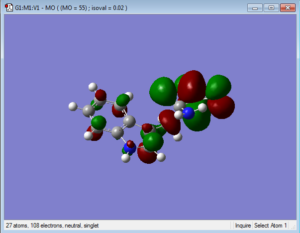
Below are representations of the HOMO and LUMO molecular orbitals of one conformation of tryptophan.